K12 Stem Exam Topic Modeling and Analysis¶
Author: Paul Shao¶
Motivation and Project Description¶
Concerns regarding the effectiveness of elementary and middle school’s science education have elevated significantly over the past few years. With a polarizing political landscape and an impasse between federal and state government over the increasing education achievement gap among students with drastically different family and financial backgrounds, an investigation of some of the most commonly asked science exam questions at elementary & middle schools across the U.S. can shed more light on how the current administration (both local and national) navigates the difficult task of providing a impartial, accurate, and modern science education while aligning with the socioeconomic and political interests of the regional voters. The goal of this project is to visualize and explore the following questions surrounding elementary STEM education in the U.S.:
- What scientific topic(s) do K-12 schools in the U.S. most commonly focus on?
- How does the emphasis on the questions vary through the descriptions of the questions across each scientific subject category?
Datasets¶
Dataset Used: To facilitate a broad understanding of how students’ understanding of different scientific subjects are assessed across K-12 schools in the U.S., I will incorporate the following dataset in my analysis:
- The Aristo Reasoning Challenge (ARC) corpus, which contains 7787 elementary and middle school standardized exam questions drawn from 12 US states.
import sys
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import torch
import sklearn
import seaborn as sns
import os
import re
import plotly.express as px
import tensorflow as tf
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
from keras.utils.np_utils import to_categorical
from tensorflow.keras.callbacks import EarlyStopping, ReduceLROnPlateau
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dropout, Dense, Embedding, LSTM, SpatialDropout1D
from sklearn.model_selection import train_test_split
from sklearn.cluster import MiniBatchKMeans
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.decomposition import PCA
from wordcloud import WordCloud, STOPWORDS, ImageColorGenerator
import nltk
from nltk.corpus import stopwords
from nltk import word_tokenize
from pyLDAvis import sklearn as sklearn_lda
import pickle
import pyLDAvis
import warnings
warnings.filterwarnings('ignore')
qc_questions = pd.read_csv('QC-ARC-Questions.tsv',delimiter='\t',encoding='utf-8')
qc_taxonomy = pd.read_csv('QC-ARC-Taxonomy.tsv', delimiter='\t', encoding='utf-8')
qc_questions.head()
qc_questions['QCLabel'].value_counts()
qc_questions.shape[0]
Exploratory Data Analysis¶
For this stage, I will be trying out a variety of visualization techniques on the K12 QC ARC question banks based on their categories and the frequencies of similar questions across different subjects.
Techniques to Include:¶
- WordCloud
- TF-IDF Vectorizer for Feature Extraction
- Clustering Using MiniBatch K-Means
WordCloud¶
To perform a WordCloud visualization on the questions, we will be running all the questions through the following data pipeline:
- tokenization
- remove stop words
- tagging
- keeping only nouns
- apply port stemmer and remove unit-length word
nltk.download('punkt')
nltk.download('averaged_perceptron_tagger')
nltk.download('stopwords')
stop_words = set(stopwords.words('english'))
def port_stem(s):
p_stemmer = nltk.PorterStemmer()
return [p_stemmer.stem(word) for word in s]
tokenized_questions = qc_questions['Question'].str.lower().apply(nltk.word_tokenize)
tokenized_questions = tokenized_questions.apply(lambda tokens: [w for w in tokens if not w in stop_words])
tokenized_q_pos = tokenized_questions.apply(nltk.pos_tag)
tokenized_nouns = tokenized_q_pos.apply(lambda tags: [word for word,pos in tags if (pos == 'NN' or pos == 'NNP' or pos == 'NNS' or pos == 'NNPS')])
tokenized_nouns = tokenized_nouns.apply(port_stem).apply(lambda nouns: [noun for noun in nouns if len(noun) > 1])
retokenized = tokenized_nouns.apply(lambda tokens: ' '.join(tokens))
qtext = ''
for token in retokenized:
qtext += token + ' '
wordcloud = WordCloud(background_color="white").generate(qtext)
plt.figure(figsize=(12, 6))
plt.imshow(wordcloud, interpolation='bilinear')
plt.axis("off")
plt.show()
category = qc_questions['QCLabel'].apply(lambda x: x.split('_')[0]).apply(lambda x: 'LIFE' if x == 'OTHER, LIFE' else x)
qc_questions['Subject'] = category
plt.title('Distribution of Subjects Per Exam Question')
sns.countplot(x='Subject', data=qc_questions, palette="Set3", order = qc_questions['Subject'].value_counts().index);
Above is a visualization of the distribution of the primary subjects of each question pre-assigned by the ARC. As we can see, Life science, Earth Science, and Material Science (along with a focus on matters and measurements in physics) predominantly represent the majority of the question bank.
TF-IDF Vectorization and MiniBatch K-Means¶
As an initial next step, to visualization how each exam question is phrased and the simillarity/difference among them across different subjects, we will compute the TF-IDF values as part of our feature extraction process and attempt to run an initial MiniBatch K-means clustering algorithm on the data.
As a first step, we will obtain the feature matrix on all the questions using the TfidfVectorizer.
REPLACE_BY_SPACE_RE = re.compile('[/(){}\[\]\|@,;]')
BAD_SYMBOLS_RE = re.compile('[^0-9a-z #+_]')
nltk.download('stopwords')
STOPWORDS = set(stopwords.words('english'))
def clean_text(text):
text = text.lower()
text = REPLACE_BY_SPACE_RE.sub(' ', text)
text = BAD_SYMBOLS_RE.sub('', text)
text = text.replace('x', '')
text = ' '.join(word for word in text.split() if word not in STOPWORDS)
return text
qc_questions['Q_cleaned'] = qc_questions['Question'].apply(clean_text).str.replace('\d+', '')
vec = TfidfVectorizer(stop_words="english")
vec.fit(qc_questions['Q_cleaned'].values)
features = vec.transform(qc_questions['Q_cleaned'].values)
In order to determine how many clusters we would like our K-means algorithm to aim for, we can start with a random guess and gradually increase the number of clusters.
cls_s = []
for i in range(5, 15):
cls = MiniBatchKMeans(n_clusters=i, random_state=0)
cls.fit(features)
cls_s.append(cls)
for cls in cls_s:
cls.predict(features)
To visualize, we’ll plot the features in a 3D space. As we know the dimension of features that we obtained from TfIdfVectorizer could be quite large, we need to reduce the dimension before we can plot. For this, we’ll ues PCA to transform our high dimensional features into 3 dimensions.
Note: As we hover over individual data points (exam questions) in the 3D scatterplot below, we can see the corresponding subject for the question.
pca = PCA(n_components=3)
reduced_features = pca.fit_transform(features.toarray())
reduced_cluster_centers = pca.transform(cls.cluster_centers_)
5 Clusters¶
import plotly.graph_objects as go
fig = go.Figure(data=[
go.Scatter3d(x=reduced_features[:,0], y=reduced_features[:,1], z=reduced_features[:,2],
mode='markers',text=qc_questions['Subject'], marker=dict(size=4,color=cls_s[0].predict(features),
colorscale='Rainbow',opacity=0.6)),
go.Scatter3d(x=reduced_cluster_centers[:,0], y=reduced_cluster_centers[:,1], z=reduced_cluster_centers[:,2],
mode='markers',marker=dict(size=6, symbol='x', color='rgb(0, 0, 0)'))
]
)
fig.update_layout(
scene = dict(
xaxis_title='PC1',
yaxis_title='PC2',
zaxis_title='PC3'
),
title='K-Means Clustering of Exam Questions (5 Clusters)'
)
fig.show()
10 Clusters¶
fig = go.Figure(data=[
go.Scatter3d(x=reduced_features[:,0], y=reduced_features[:,1], z=reduced_features[:,2],
mode='markers',text=qc_questions['Subject'], marker=dict(size=4,color=cls_s[5].predict(features),
colorscale='Rainbow',opacity=0.6)),
go.Scatter3d(x=reduced_cluster_centers[:,0], y=reduced_cluster_centers[:,1], z=reduced_cluster_centers[:,2],
mode='markers',marker=dict(size=6, symbol='x', color='rgb(0, 0, 0)'))
]
)
fig.update_layout(
scene = dict(
xaxis_title='PC1',
yaxis_title='PC2',
zaxis_title='PC3'
),
title='K-Means Clustering of Exam Questions (10 Clusters)'
)
fig.show()
15 clusters¶
fig = go.Figure(data=[
go.Scatter3d(x=reduced_features[:,0], y=reduced_features[:,1], z=reduced_features[:,2],
mode='markers', text=qc_questions['Subject'], marker=dict(size=4,color=cls_s[-1].predict(features),
colorscale='Rainbow',opacity=0.6)),
go.Scatter3d(x=reduced_cluster_centers[:,0], y=reduced_cluster_centers[:,1], z=reduced_cluster_centers[:,2],
mode='markers',marker=dict(size=6, symbol='x', color='rgb(0, 0, 0)'))
]
)
fig.update_layout(
scene = dict(
xaxis_title='PC1',
yaxis_title='PC2',
zaxis_title='PC3'
),
title='K-Means Clustering of Exam Questions (15 Clusters)'
)
fig.show()
As we can see above, with a pre-determined set of 15 clusters and a principle component analysis (PCA) in 3 dimensions, we can see that the topics are broken down along mainly 3 categories: Engineering, Cell Biology, and Earth/Material Science. Toward the center, the topics are concentrated on Forensics and Life Science.
LDA (Latent Dirichlet Allocation)¶
LDA (Latent Dirichlet Allocation) is a generative statistical model that used unsupervised learning to group texts together by extracting the underlying topics and comparing their levels of similarity and marginal frequencies. Here, when applying a pre- determined 20 topics, we can see that the topics are further divided into the following main subjects:
- Life Science (Organisms)
- Earth Science (Ecology)
- Measurement (Experiments)
- Biology (Cells and Chemistry)
- Earth Science (Geology)
- Physics (Energy and Matters)
- Miscellaneous (Students)
from sklearn.decomposition import LatentDirichletAllocation as LDA
from sklearn.feature_extraction.text import CountVectorizer
count_vectorizer = CountVectorizer(stop_words='english')
count_data = count_vectorizer.fit_transform(qc_questions['Question'])
def count_topics(model, count_vectorizer, n_top_words):
words = count_vectorizer.get_feature_names()
for topic_idx, topic in enumerate(model.components_):
print("\nTopic #%d:" % topic_idx)
print(" ".join([words[i]
for i in topic.argsort()[:-n_top_words - 1:-1]]))
number_topics = 20
number_words = 10
lda = LDA(n_components=number_topics, n_jobs=-1)
lda.fit(count_data)
print("Topics found via LDA:")
count_topics(lda, count_vectorizer, number_words)
Visualizing the LDA Topic Modeling Results¶
LDAvis_data_filepath = 'ldavis_prepared_'+str(number_topics)
LDAvis_prepared = sklearn_lda.prepare(lda, count_data, count_vectorizer)
pyLDAvis.save_html(LDAvis_prepared, 'ldavis_prepared_'+ str(number_topics) +'.html')
pyLDAvis.display(LDAvis_prepared)
The Intertopic Distance Map above represents the similarity between each topical cluster by computing the Jensen- Shannon divergence and applying PCA to project their distances onto a 2D plane for ease of visualization.
$\lambda$ here represents the relevance of a word. A term relevance is determined by their topic-specific probability: the marginal probability the specific term appears within the given topical cluster.
LSTM-RNN Modeling¶
An LSTM-based RNN is a state-of-the-art (SOTA) deep learning architecture that performs well in predicting time-series data and working with sequence models.
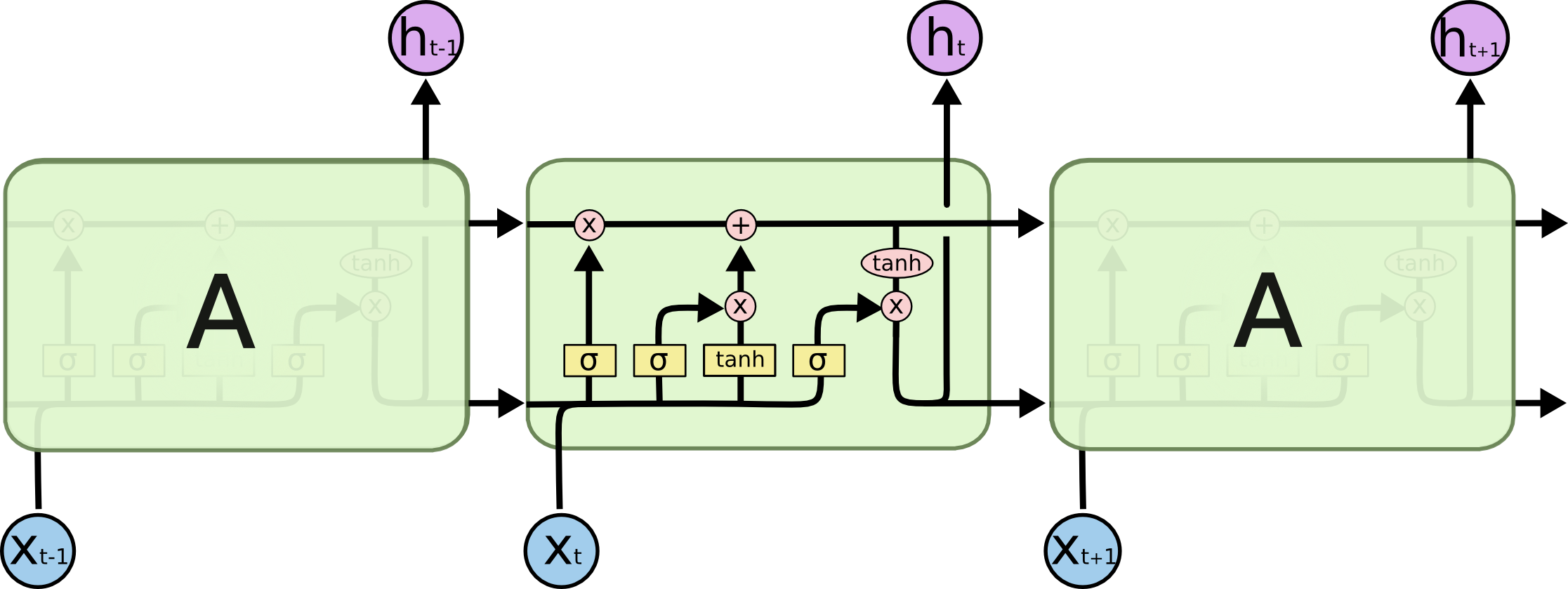
Here, an LTSM-based RNN is trained on a TF- IDF vectorized matrix containing tokens of the ACR Exam questions to predict the underlying subject (one-hot encoded) of the question. Overall a total of 10 training epochs using the Adams optimizer, we can achieve ~76% test accuracy.
import re
import nltk
from nltk.corpus import stopwords
from nltk import word_tokenize
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
from keras.models import Sequential
from keras.layers import Dense, Embedding, LSTM, SpatialDropout1D
from sklearn.model_selection import train_test_split
from keras.utils.np_utils import to_categorical
from keras.callbacks import EarlyStopping
from keras.layers import Dropout
MAX_WORD_COUNT = 5000
MAX_SEQ_SIZE = 50
EMBEDDING_DIM = 50
EPOCHS = 10
BATCH_SIZE = 32
category = qc_questions['QCLabel'].apply(lambda x: x.split('_')[0]).apply(lambda x: 'LIFE' if x == 'OTHER, LIFE' else x)
qc_questions['Subject'] = category
REPLACE_BY_SPACE_RE = re.compile('[/(){}\[\]\|@,;]')
BAD_SYMBOLS_RE = re.compile('[^0-9a-z #+_]')
nltk.download('stopwords')
STOPWORDS = set(stopwords.words('english'))
def clean_text(text):
text = text.lower()
text = REPLACE_BY_SPACE_RE.sub(' ', text)
text = BAD_SYMBOLS_RE.sub('', text)
text = text.replace('x', '')
text = ' '.join(word for word in text.split() if word not in STOPWORDS)
return text
qc_questions['Q_cleaned'] = qc_questions['Question'].apply(clean_text).str.replace('\d+', '')
tokenizer = Tokenizer(num_words=MAX_WORD_COUNT, filters='!"#$%&()*+,-./:;<=>?@[\]^_`{|}~', lower=True)
tokenizer.fit_on_texts(qc_questions['Q_cleaned'].values)
word_index = tokenizer.word_index
print('Found %s unique tokens.' % len(word_index))
X = tokenizer.texts_to_sequences(qc_questions['Q_cleaned'].values)
X = pad_sequences(X, maxlen=MAX_SEQ_SIZE)
print('Shape of Design Matrix:', X.shape)
y = pd.get_dummies(qc_questions['Subject']).values
print('Shape of Response Vector:', y.shape)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.15, random_state = 42)
print(X_train.shape,y_train.shape)
print(X_test.shape,y_test.shape)
model = Sequential()
model.add(Embedding(MAX_WORD_COUNT, EMBEDDING_DIM, input_length=X.shape[1]))
model.add(LSTM(50))
model.add(Dense(9, activation='relu'))
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
print(model.summary())
history = model.fit(X_train, y_train, epochs=EPOCHS, batch_size=BATCH_SIZE, validation_split=0.1)
accr = model.evaluate(X_test,y_test)
print('Test set\n Loss: {:0.3f}\n Accuracy: {:0.3f}'.format(accr[0],accr[1]))
plt.title('Loss')
plt.plot(history.history['loss'], label='train')
plt.plot(history.history['val_loss'], label='test')
plt.legend()
plt.show()
plt.title('Accuracy')
plt.plot(history.history['accuracy'], label='train')
plt.plot(history.history['val_accuracy'], label='test')
plt.legend()
plt.show();